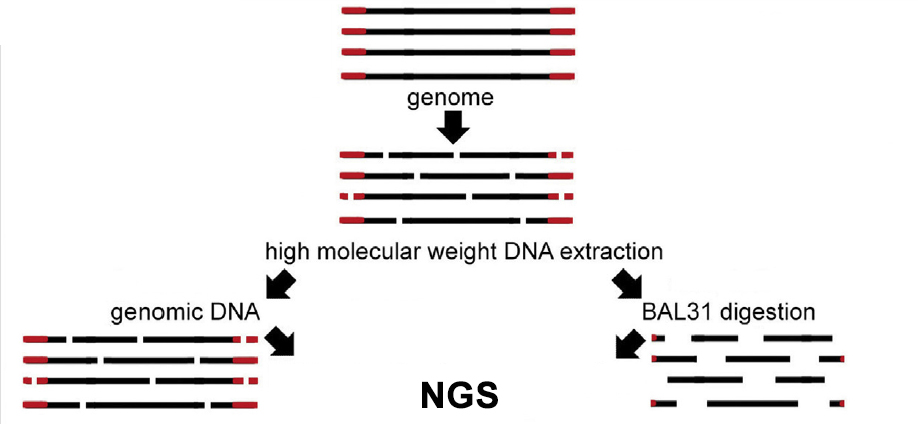
Identification of unknown telomere DNA is based on our original BAL31-NGS approach (Peška et al. Methods 2017). The protocol starts with isolation of high molecular weight DNA which is split into two aliquots. One of these aliquots is then treated with BAL31 nuclease which progressively digests the ends of linear DNA fragments. Each of aliquots is then used for preparation of a library for NGS and results obtained with both libraries are compared. Repeated sequences which are relatively depleted in a library obtained from BAL31-treated DNA are identified and their bona fide telomeric role is demonstrated using fluorescence in situ hybridisation, BAL31-sensitivity of terminal restriction fragments and TRAP assay with sequencing of its products.
This technology can be used for:
Identification of DNA sequences functioning as telomeric DNA is needed for chromosome engineering and telomere-associated truncation of chromosomes in plants and animals.
Equipment
- Next Generation Sequencing instrumentation
- Data storage and processing equipment
- RepeatExplorer and Tandem Repeats Finder tools (see Peška et al., Methods 2017)


 Share
Share
