WALTER
WALTER (Web-based Analyser of the Length of Telomeres) is a web-based toolset for terminal restriction fragments (TRF) scan analysis. It is consisted of two tools: ScanToIntensity and IntensityAnalyser. ScanToIntensity tool is able to convert selected areas of TRF scan into intensity profiles while IntensityAnalyser tool analyses the resultant file from ScanToIntensity to provide a boxplot or a violin plot depiction of said profiles with a possible statistical analysis.

WALTER toolset is running on the servers of Core Facility Bioinformatics, CEITEC. Toolset development was supported by the Czech Science Foundation, project no. 18-07027S.
In case you have used our toolset, please do cite:
Lyčka, M., Peska, V., Demko, M. et al. WALTER: an easy way to online evaluate telomere lengths from terminal restriction fragment analysis. BMC Bioinformatics 22, 145 (2021). https://doi.org/10.1186/s12859-021-04064-0
Links to tools:
Direct connection (calculations might take longer in case more people are using the toolset at the same time due to the waiting for other users' process to finish):
- ScanToIntensity tool - tool that allows the transformation of the picture to intensity profiles
- IntensityAnalyser tool - tool that can analyze a file with intensity profiles
Servers might be occasionally under maintenance which will make the toolset unavailable. In such occasion, we do recommend users to download the portable version.
Portable version of the WALTER toolset (Windows OS only):
- Download WALTER toolset v2.1 Portable
- Quick guide how to run the portable version or how to run the toolset locally on other OS
If you are not able to access the toolset or have any other problems, suggestions or a feedback, please contact us on the e-mail: 408297@mail.muni.cz
- Example of the analysis by WALTER - .zip file with TRF scan and subsequent analysis for learning purposes
- WALTER toolset v2.0 User Manual
- GitHub repository (WALTER source code, README file, license)
Update information:
14/02/2025
WALTER toolset has been transferred to a new server.
08/02/2025
WALTER toolset v2.1 Portable was released. The new version of portable toolset solves compatibility issue with Java that was responsible for sudden crashes of both tools.
22/03/2021
WALTER toolset has been published in the journal BMC Bioinformatics:
12/12/2020
WALTER toolset v2.0 launched (ScanToIntensity output from v1.0 is not compatible with IntensityAnalyser v2.0)
11/05/2020
WALTER toolset v1.0 launched
TeloBase
TeloBase is a curated database containing computationally predicted and experimentally confirmed telomere sequences of species across the whole tree of life.
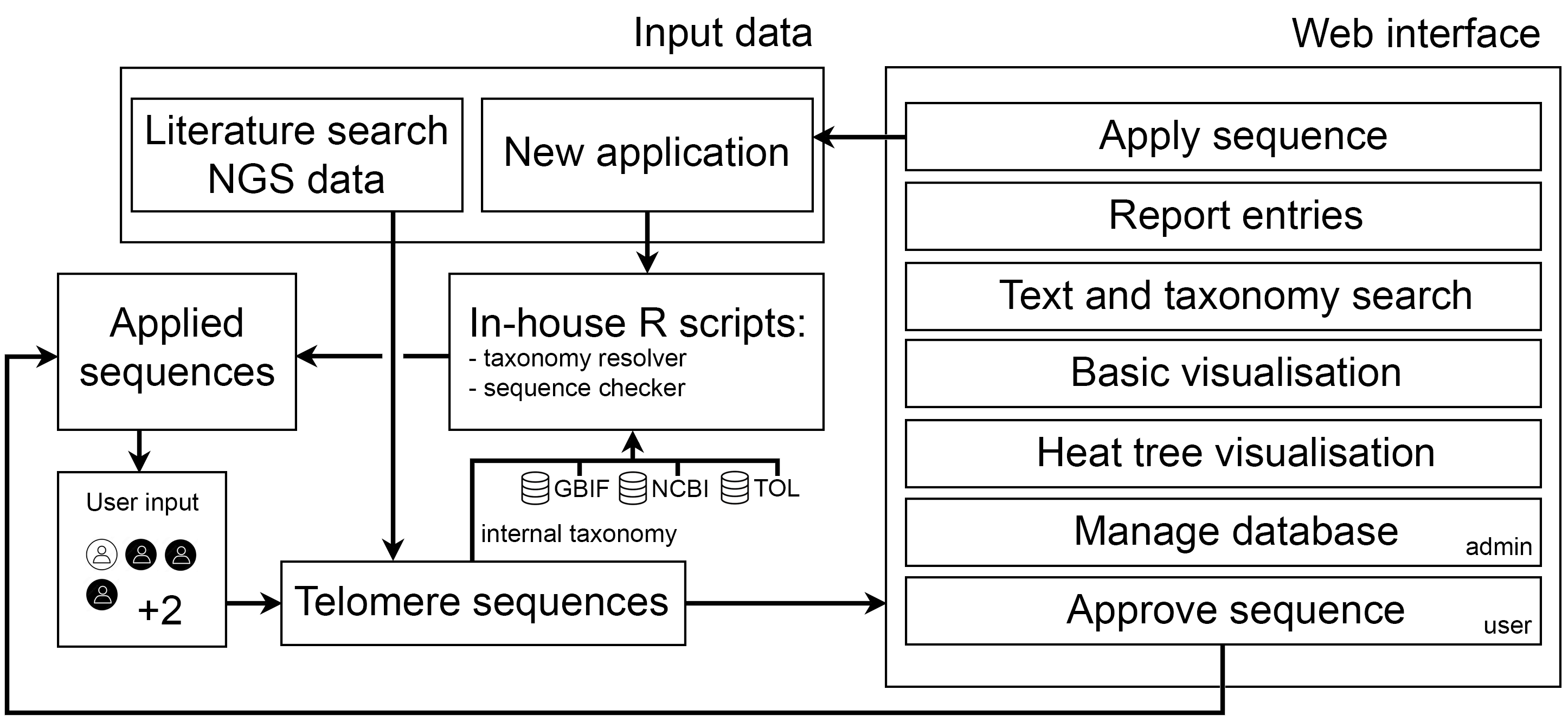
TeloBase is running on the servers of Core Facility Bioinformatics of CEITEC Masaryk University, Brno.
Development was supported by the Czech Science Foundation, project 20-01331X, and Ministry of Education, Youth and Sports of the Czech Republic, INTER-COST project LTC20003.
In case you have used our toolset, please do cite:
Martin Lyčka, Michal Bubeník, Michal Závodník, Vratislav Peska, Petr Fajkus, Martin Demko, Jiří Fajkus, Miloslava Fojtová, TeloBase: a community-curated database of telomere sequences across the tree of life, Nucleic Acids Research, 2023;, gkad672, https://doi.org/10.1093/nar/gkad672
Link to the database:
Servers might be occasionally under maintenance which will make the database temporarily unavailable.
If you are not able to access the database, please contact us on the e-mail: 408297@mail.muni.cz
- TeloBase User Manual
- raw TRFi data - .xlsx file of data from TRFi pipeline with highlighted sequences of predicted telomere motifs for species implemented in TeloBase
- Atlas of telomere sequences - an atlas showing telomere DNA evolutionary dynamics currently known from literature (until September 2022) in form of heat tree plots
Update information:
14/02/2025
TeloBase has been transferred to a new server. The previous URL temporarily redirects to the new server address. Please note that the old link may work for a limited time.
21/08/2023
TeloBase has been published in the journal Nucleic Acids Research:
TeloBase: a community-curated database of telomere sequences across the tree of life
01/02/2023
TeloBase launched




