29. Jan. 2021
Press release;
A team of scientists, led by professor Martin Lysak from CEITEC MU, revealed that the origin of a monophyletic plant clade could be more complex than previously thought. The researchers discovered that the contentious relationships of the enigmatic clade Biscutelleae within the mustard family arose from multiple independent hybridization and whole-genome duplication events involving the same and/or closely related ancestral genomes. Their findings have provided new insights into the diversification and evolution of plant lineages. The paper describing their findings was published in December 2020 in the scientific journal Molecular Biology and Evolution.
Polyploidy or whole-genome duplication (WGD) is an event forming an organism with additional copies of the genome. Whereas autopolyploids originate by crossing between genetically near-identical individuals, allopolyploid genomes are formed through inter-species crosses, combining two or more different genomes. After the polyploidization, complex genomic processes collectively called diploidization usually revert polyploid genomes back to functionally diploid ones. Recurrent hybridization, polyploidization and diploidization on timescales over millions of years may create polyploid complexes comprising diploidizing polyploids of a different age with contentious phylogenetic relationships.
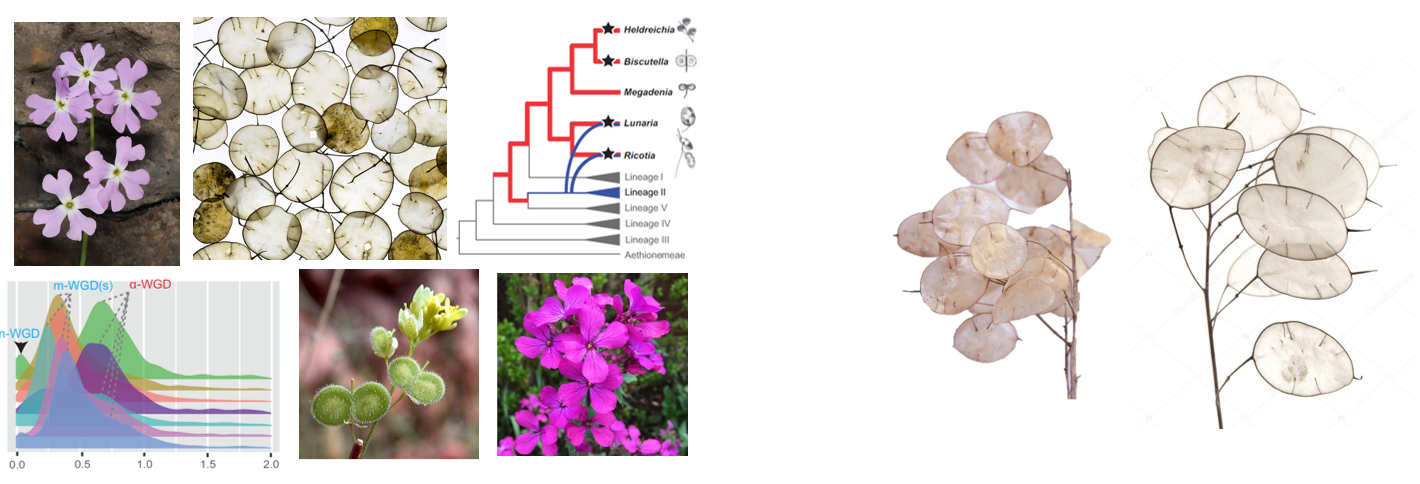
“If all extant representatives of a plant lineage are considered to have a polyploid origin, scientists tend to infer that the group originated from a single whole-genome duplication event. However, we have realized that such a conclusion seems not to fit the evolutionary history of one of the lineages, called the Biscutelleae, we are studying in the group”, explained corresponding author Martin Lysak. Researchers in the Martin Lysak research group previously found that two genera of the Biscutelleae clade (Biscutella and Ricotia) had a polyploid origin but the structure of their putative parental genomes appeared to differ. Together with the puzzling phylogenetic placement of Biscutelleae species, these findings prompted the researchers to reconstruct genome evolution in all five recognized genera of this clade.
The structures of these genomes were elegantly reconstructed by Terezie Mandáková, the co-first author of the study, using comparative chromosome painting. Based on the similarity of genomic sequences among species of the mustard family, fluorescently labelled DNA fragments derived from the Arabidopsis thaliana genome were “hybridized” to chromosomes of the Biscutelleae species to visualize the arrangement of targeted genomic regions. The researchers observed that only a single genus (Megadenia) remained diploid, while the four remaining genera originated by independent allopolyploid (Biscutella, Lunaria, Ricotia) or autopolyploid (Heldreichia) genome duplications. They also described a number of chromosome rearrangements which occurred during post-polyploid diploidization of these genomes, including the three times repeated independent origin of one chromosome from the same ancestral ones.
“But having the genome structures of these species cannot provide us with sufficient confidence in their relationships, we needed complementary evidence from a phylogenomic perspective”, added Xinyi Guo, a postdoc in the Lysak group and the co-first author of the study. Through transcriptome sequencing, the researchers selected multiple sets of phylogenetic markers, including both single-copy and duplicated genes, to reconstruct the evolutionary relationships among the species and to infer the mode of their WGDs. While they confirmed the monophyly of Biscutelleae, they also corroborated the existence of several independent WGDs in the lineage. They found that the contentious relationships among Biscutelleae genera could be reflected by phylogenetic incongruence throughout the alignable regions of their coding sequences.
“By using complementary cytogenomics and phylogenomics approaches, we were able to show that the origin of a monophyletic plant clade can be more complex than a simple and commonly used assumption that a single whole-genome duplication preceded the divergence of a plant lineage. Instead, recurrent hybridization among the same and/or closely related parental genomes may phylogenetically interlink independently formed polyploid genomes”, concluded Lysak. In the future, the team would like to further untangle the evolutionary consequences of genome duplications of different ages and involving more versus less related parental genomes.
Author


 Share
Share